DeepPhysioRecon
Reconstructing physiological signals from fMRI dynamics
Many fMRI studies lack physiological measurements, which limits both the interpretation and richness of the data. Natural fluctuations in breathing and heart rate provide windows into cognition, emotion, and health—and they also heavily influence fMRI signals. DeepPhysioRecon addresses this gap by decoding continuous variations in respiration and heart rate directly from whole-brain fMRI dynamics.
The Problem
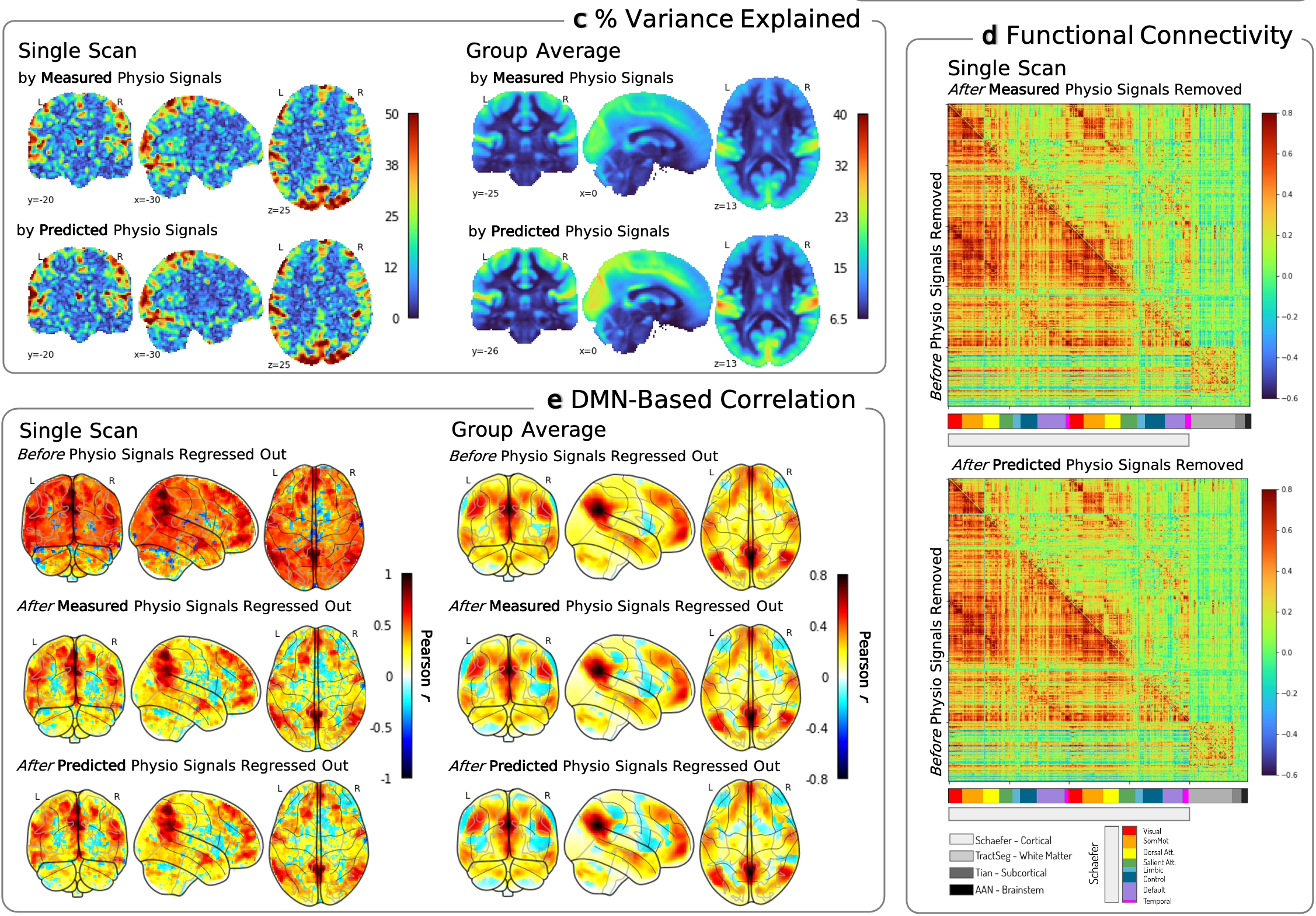
The brain and body are closely coupled. Functional MRI measures blood oxygenation, which is influenced not only by neural activity but also by physiological processes like breathing and heart rate. These low-frequency fluctuations (below 0.15 Hz) can account for substantial variance in fMRI signals—in some brain regions, up to 50% of the signal.
However, many existing fMRI datasets lack physiological recordings entirely, and even when recordings exist, they are often noisy or corrupted. This limits our ability to properly interpret fMRI results or study brain-body interactions.
Our Approach
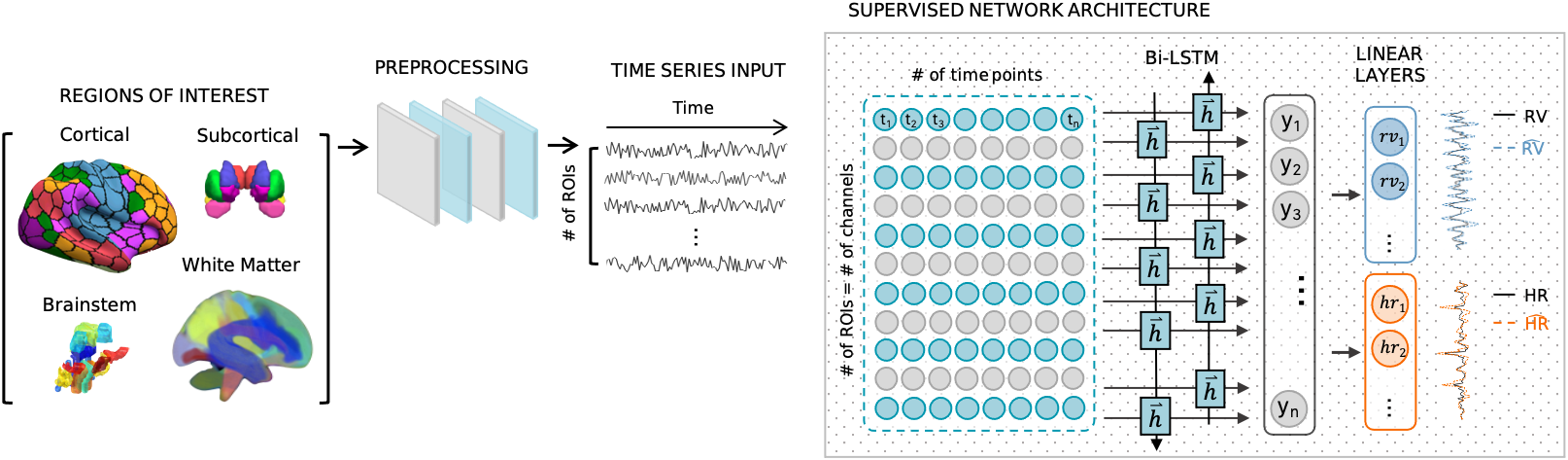
DeepPhysioRecon uses a bidirectional Long Short-Term Memory (LSTM) network to jointly predict respiration variation and heart rate from fMRI time-series data. The key insight is that RV and HR are coupled, so learning them together improves accuracy for both.
The framework works with conventional fMRI acquisitions (TR > 1s) and does not require fast multiband sampling or slice-based reconstructions. This makes it applicable to the vast majority of existing fMRI datasets.
Key Results
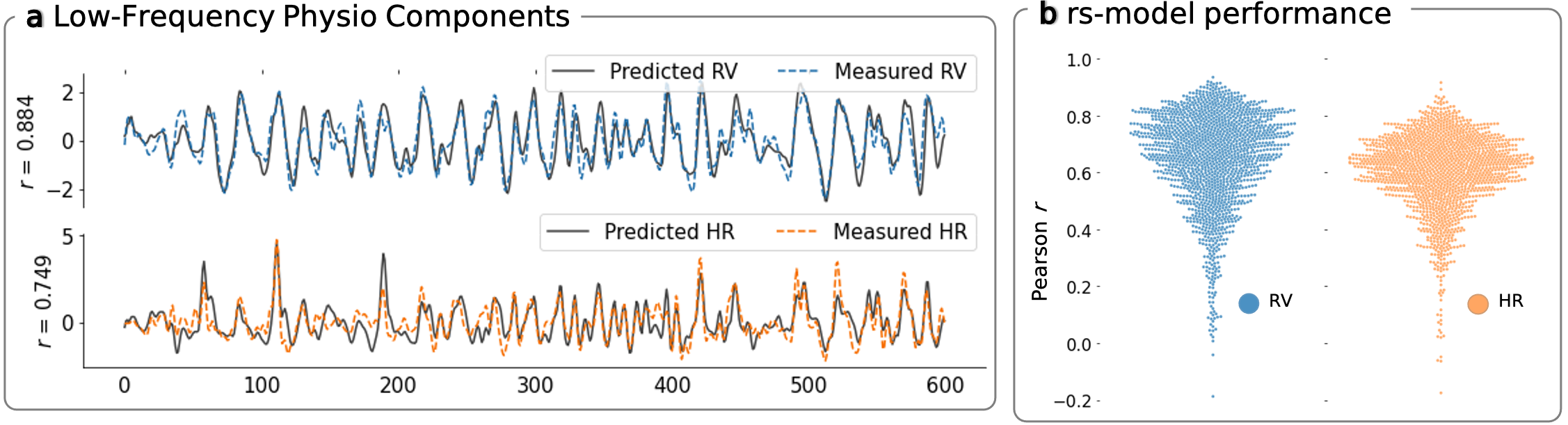
Models trained on resting-state fMRI data from the Human Connectome Project achieved median correlations of approximately 0.68 for RV and 0.63 for HR against measured physiological signals. The models generalized across experimental conditions, different subjects, and even different scanners and acquisition parameters.
The predicted signals can be used to account for physiological effects in fMRI analyses. We showed that removing predicted RV and HR from fMRI data alters functional connectivity maps in ways that closely match what happens when using measured physiological signals.
Generalizability
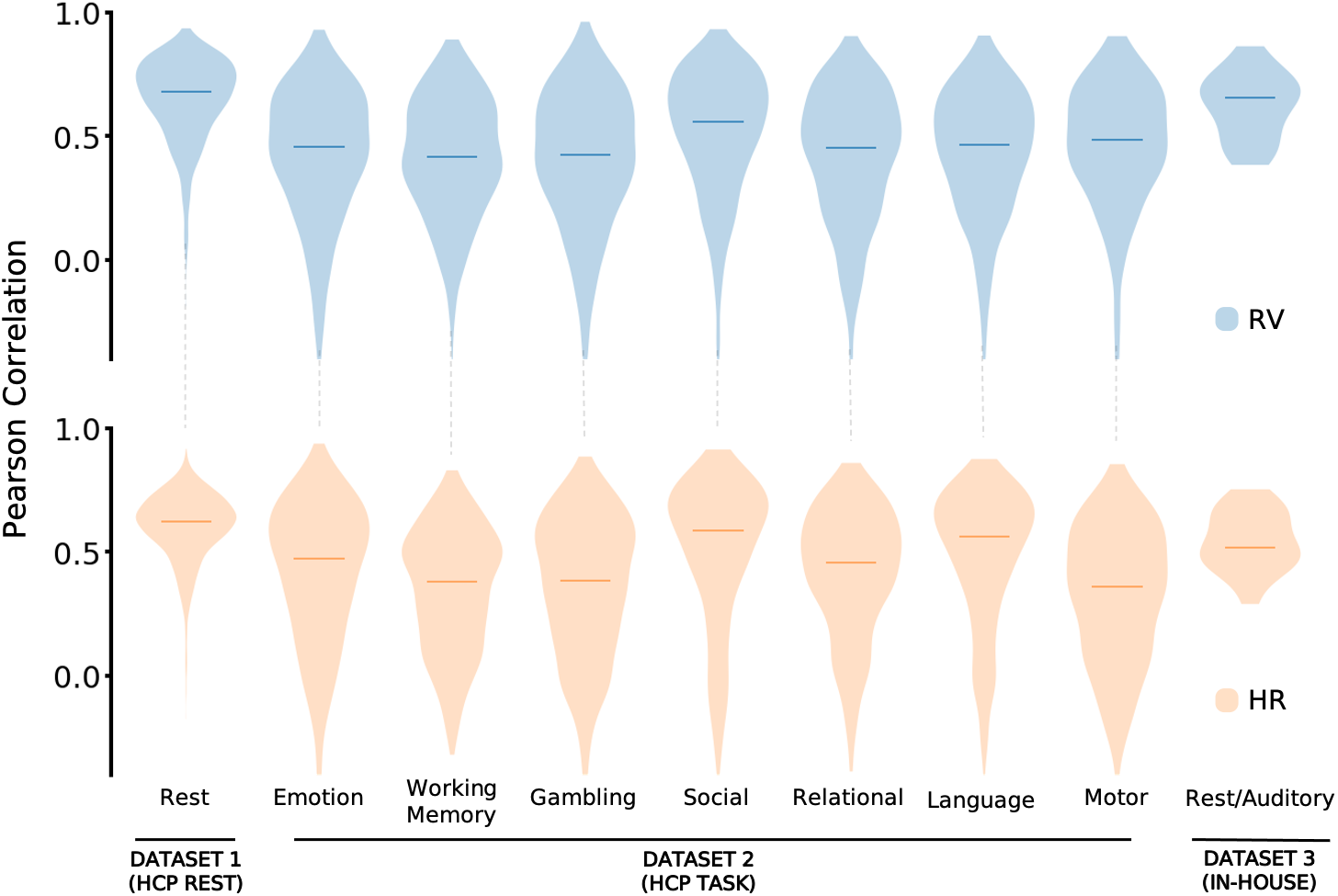
We tested the models across seven different task conditions from HCP and an external dataset with different acquisition parameters (including TR of 2.1s vs. 0.72s). The resting-state trained models transferred well to task data, and task-trained models showed improved performance on held-out tasks.
This suggests that the relationship between fMRI and physiology is largely brain state-invariant, making pre-trained models broadly applicable.
Potential for Denoising
An unexpected finding: in cases where measured physiological recordings contained artifacts, the predicted signals appeared to smooth over these corruptions. This suggests DeepPhysioRecon may also be useful for cleaning up noisy physiological data, not just replacing missing data.
Code and Data
DeepPhysioRecon is open source and available for use:
- Code: GitHub
- Model weights: Hugging Face
- Paper: Imaging Neuroscience
Citation
Bayrak RG, Hansen CB, Salas JA, Ahmed N, Lyu I, Mather M, Huo Y, Chang C. DeepPhysioRecon: Tracing peripheral physiology in low frequency fMRI dynamics. Imaging Neuroscience. 2025;3. 10.1162/IMAG.a.163