TractEM
Evaluating reproducibility of white matter tractography segmentation protocols
TractEM is an open, public protocol for segmenting white matter pathways from diffusion MRI tractography data. This project evaluated the reproducibility of manual bundle segmentation — a foundational step for building white matter atlases and understanding brain connectivity.
The Problem
Reproducible identification of white matter pathways across subjects is essential for studying brain connectivity. However, two key challenges persist: anatomical differences between individuals and human subjectivity in labeling. Existing white matter atlases often lack detailed protocols explaining how structures were delineated, making it difficult to replicate or extend their work.
A segmentation protocol is only useful if it can be followed consistently—both by the same person over time (intra-rater reliability) and by different people (inter-rater reliability). Without quantifying this reproducibility, we cannot confidently interpret differences observed across subjects or studies.
Approach
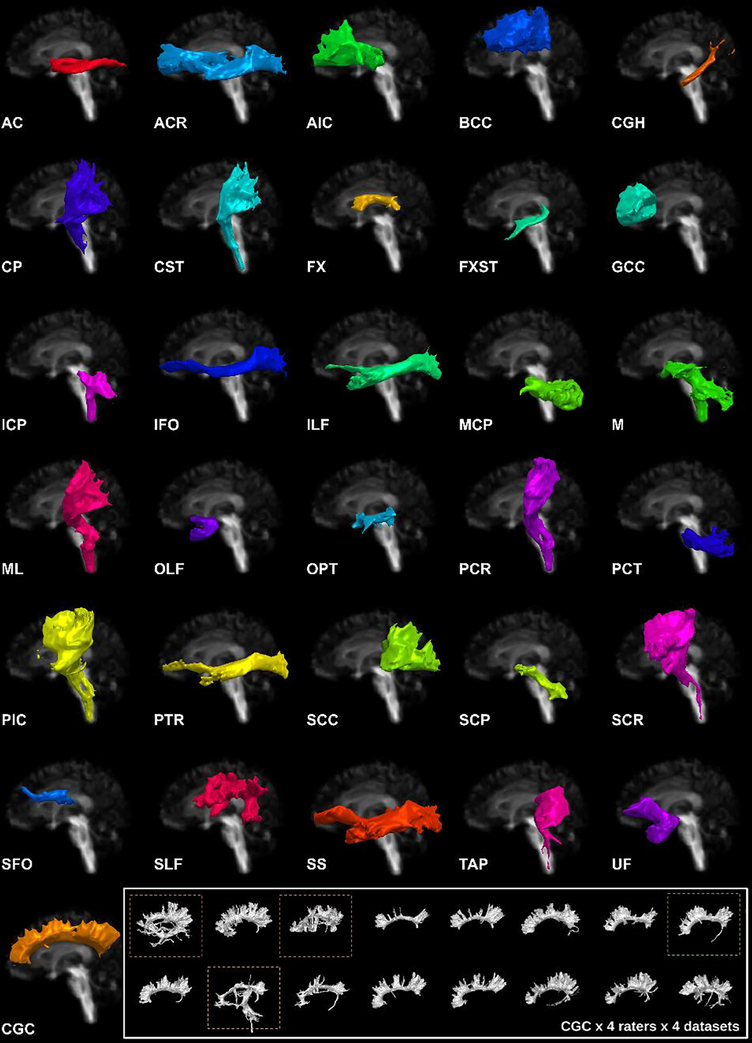
We developed TractEM as a first iteration of a whole-brain segmentation protocol informed by the EVE atlas. The protocol covers 53 unique pathways across commissural, association, projection, and brainstem bundles.
To evaluate reproducibility, we recruited raters with no prior tractography experience and had them segment bundles using DSI-Studio on two datasets: the Baltimore Longitudinal Study of Aging (BLSA) and the Human Connectome Project (HCP).
Key Findings
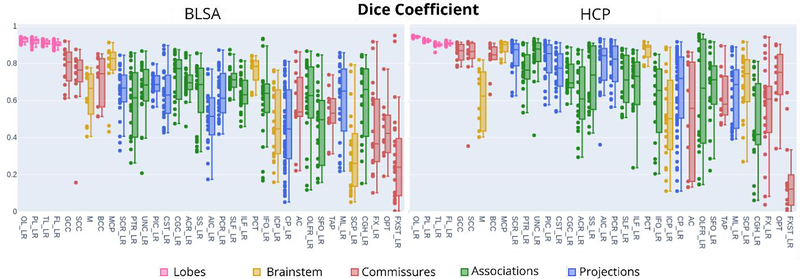
From approximately 2,600 manually identified bundles across 10 raters, we found:
Good core agreement: Most bundles achieved acceptable reproducibility in their densest regions, as measured by weighted Dice coefficient (>0.8 for BLSA, >0.85 for HCP).
Outliers remain problematic: Spurious streamlines extending beyond the bundle core negatively affected standard Dice scores and bundle adjacency metrics.
Data quality had limited impact: Contrary to our hypothesis, the higher-quality HCP data did not consistently yield better reproducibility than BLSA. Only 11 of 35 bundles showed statistically significant differences between datasets.
Protocol complexity matters: While simple bundles (single seed region) did not consistently outperform complex bundles (multiple ROIs), the number and clarity of region-of-interest definitions influenced variability.
Why It Matters
This work highlights a critical point: not only must the neuroimaging community reach consensus on anatomical definitions of white matter pathways, but the interpretation and execution of those definitions must be quantified to assess practical utility.
The TractEM protocol and all data have been made openly available to facilitate collaboration and iterative improvement. This baseline evaluation identifies specific limitations—such as unclear ROI descriptions and lack of visual guidance—that can be addressed in future versions.
Resources
The protocol, raw data, and results are available at the TractEM project website.
Citation: Rheault F, Bayrak RG, Wang X, et al. TractEM: Evaluation of protocols for deterministic tractography white matter atlas. Magnetic Resonance Imaging. 2022;85:44-56. doi:10.1016/j.mri.2021.10.017